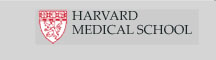ABOUT
The Core
 The NextGen Core was founded as a collaboration between the Molecular Biology, Center for Human Genetics Research, and Center for Computational Biology departments.
The NextGen Core was founded as a collaboration between the Molecular Biology, Center for Human Genetics Research, and Center for Computational Biology departments.
Currently, the Core operates using a single Ilumina HiSeq instrument, accompanied by Illumina's cBot for cluster generation. This upgrade from our Genome Analyzer II doubled our capacity and greatly increased the data amount, quality, and stability over extra-long reads.
The Core is located in the state-of-the-art Richard Simches Research Center on Cambridge St. as part of the MGH main campus in Boston. The many multi-investigator groups in the building - including those that study human genetics, stem cells, genomics, and more - make it the perfect location for the Core to service the researchers in those groups. The majority of customers come from MGH, but we also service customers at other academic medical centers and industry.
Applications
There is a wide array of sequencing applications, some of which are well-established, others of which are novel and developed by researchers on a project-specific basis.
Some of these applications are:
• Genomic sequencing or resequencing
• Chromatin-IP analysis (ChIP-Seq)
• Transciptome analysis (RNA-Seq)
• sRNA and miRNA discovery and profiling
• Polymorphism detection, including SNPs, genomic rearrangements, and CNVs
• Gene expression analysis
• Methylation analysis
To find out more information and learn how sequencing can be applied to your project, please contact us.
Sequencing Technology
The Sequencing Core Facility at MGH uses the Illumina HiSeq platform, a cutting-edge technology that can produce 100 million reads or more per lane. It works by binding randomly fragmented DNA to an optically-transparent surface called a flowcell, which has 8 separately-contained lanes, allowing for the sequencing of 8 samples simultaneously. Additionally, samples can be barcoded, permitting analysis of multiple samples per lane. Illumina's new TruSeq line of sample prep kits allow barcoding of up to 12 DNA or RNA samples in one lane and up to 48 small RNA samples in one lane.
Each template fragment is amplified via bridge amplification to generate millions of clusters, with each cluster containing 1,000 copies of the template. The clusters are sequenced one base at a time using sequencing-by-synthesis chemistry and laser-pwered imaging of reversible terminator, fluorescently-tagged bases (see diagram to the left).

As mentioned above, each lane yields 90-100 million+ reads, depending on the sample, and each read may be sequenced either 50 or 100 cycles at each end, with greater numbers of longer reads very possible for the near future. High-quality, low error-rate data is generated quickly, and the low cost per base makes it a very valuable tool for the everyday lab sequencing DNA and RNA from various sources.
For a complete explanation of Illumina's technology, please visit Illumina's website.