RESOURCES
Overview of Illumina Chemistry
Regardless of the library construction method, submitted libraries will consist of a sequence of interest flanked on either side by adapter constructs. On each end, these adapter constructs have flow cell binding sites, P5 and P7, which allow the library fragment to attach to the flow cell surface. The constructs also contain several other primer binding sites which are described further below. Before attachment to the flowcell, the library fragments are denatured, and thus a single-stranded copy of the library fragment is sequenced.
The diagrams below show the basic chemistry steps that occur during each read:
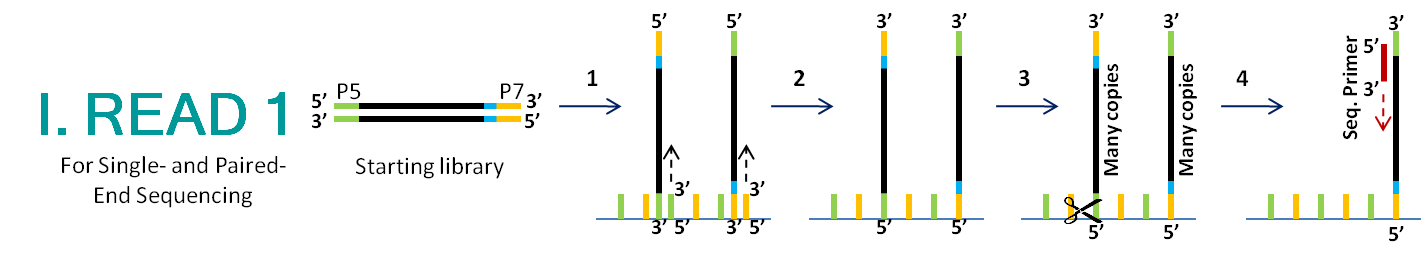
- The P5 and P7 regions of single-stranded library fragments anneal to their complimentary oligos on the flowcell surface. The flow cell oligos act as primers and a strand complimentary to the library fragment is synthesized.
- The original strand is washed away, leaving behind fragment copies that are covalently bonded to the flowcell surface in a mixture of orientations.
- 1,000 copies of each fragment are generated by bridge amplification, creating clusters. For simplification, the diagram shows only one copy (out of 1,000) in each cluster, and only two clusters (out of 30-50 million).
- The P5 region is cleaved, resulting in clusters containing only fragments which are attached by the P7 region. This ensures that all copies are sequenced in the same direction. The sequencing primer anneals to the P5 end of the fragment, and begins the sequencing by synthesis process.
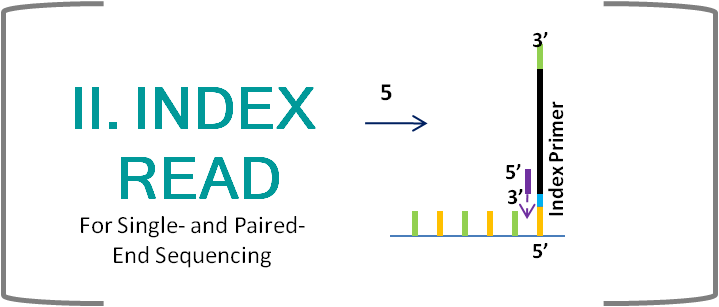
- Index reads are only performed when a sample is barcoded. When Read 1 is finished, everything from Read 1 is removed and an index primer is added, which anneals at the P7 end of the fragment and sequences the barcode.
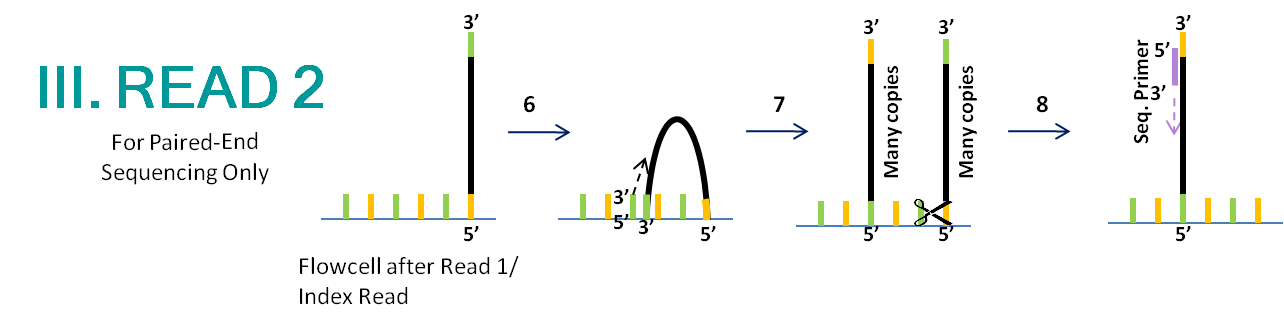
- Everything is stripped from the template, which forms clusters by bridge amplification as in Read 1. This leaves behind fragment copies that are covalently bonded to the flowcell surface in a mixture of orientations.
- This time, P7 is cut instead of P5, resulting in clusters containing only fragments which are attached by the P5 region. This ensures that all copies are sequenced in the same direction (opposite Read 1).
- The sequencing primer anneals to the P7 region and sequences the other end of the template.